欢迎大家关注全网生信学习者系列:
- WX公zhong号:生信学习者
- Xiao hong书:生信学习者
- 知hu:生信学习者
- CDSN:生信学习者2
介绍
微生物短链脂肪酸(SCFAs)是由肠道微生物发酵膳食纤维、抗性淀粉、低聚糖等碳水化合物产生的一类有机脂肪酸,主要包括乙酸、丙酸和丁酸。SCFAs在肠道健康、免疫调节、能量代谢等方面发挥重要作用,并且与多种疾病状态有关。
通过约束线性混合效应模型(CLME)研究SCFA与排序暴露分组之间的单调递增关系,这种方法通常用于分析具有层次结构的数据,能够处理数据中的非独立性和非正态分布问题。在研究SCFA与某些因素(如饮食、疾病状态等)的关系时,CLME可以用来评估这些因素对SCFA水平的影响,并检测它们之间是否存在单调递增或递减的关系。
加载R包
library(readr)
library(tidyverse)
library(CLME)
library(magrittr)
library(qwraps2)
library(ggprism)
library(ggsci)
library(ggpubr)
library(rstatix)
library(jtools)
library(kableExtra)
导入数据
大家通过以下链接下载数据:
- 百度网盘链接:https://pan.baidu.com/s/1fz5tWy4mpJ7nd6C260abtg
- 提取码: 请关注WX公zhong号_生信学习者_后台发送 复现gmsb 获取提取码
df_merge <- read_csv("./data/GMSB-data/df_merge.csv", show_col_types = FALSE)head(df_merge[, 1:6])
| sampleid | subjid | visit | visit_num | status | age |
|---|---|---|---|---|---|
| F-1 | DUMMY1 | v1 | 10 | nc | 58 |
| F-2 | DUMMY2 | v1 | 10 | nc | 60 |
| F-3 | DUMMY3 | v1 | 10 | nc | 58 |
| F-4 | DUMMY4 | v1 | 10 | nc | 72 |
| F-5 | DUMMY5 | v1 | 10 | nc | 57 |
| F-6 | DUMMY6 | v1 | 10 | nc | 59 |
数据预处理
对不同visit进行分组中,提取它们的SCFA短链脂肪酸
df_v1 <- df_merge %>%dplyr::filter(visit == "v1",group1 != "missing")
df_v2 <- df_merge %>%dplyr::filter(visit == "v2",group1 != "missing")df_v1$group1 <- recode(df_v1$group1,`g1` = "G1", `g2` = "G2",`g3` = "G3", `g4` = "G4")
df_v1$group2 <- recode(df_v1$group2,`g1` = "G1", `g2` = "G2",`g3` = "G3", `g4` = "G4", `g5` = "G5")df_v1 <- df_v1 %>%dplyr::transmute(subjid, status, leu3p, leu2p, recept_anal, abx_use, group1, group2, acetate_v1 = acetate, propionate_v1 = propionate, butyrate_v1 = butyrate, valerate_v1 = valerate)
df_v2 <- df_v2 %>%dplyr::transmute(subjid, acetate_v2 = acetate, propionate_v2 = propionate, butyrate_v2 = butyrate, valerate_v2 = valerate) df_v1 <- df_v1 %>%dplyr::left_join(df_v2, by = "subjid")df_v1$group1 <- factor(df_v1$group1, levels = c("G1", "G2", "G3", "G4"), ordered = TRUE)
df_v1$group2 <- factor(df_v1$group2, levels = c("G1", "G2", "G3", "G4", "G5"), ordered = TRUE)head(df_v1[, 1:6])
| subjid | status | leu3p | leu2p | recept_anal | abx_use |
|---|---|---|---|---|---|
| DUMMY1 | nc | 40.5 | 35.9 | 5 | yes |
| DUMMY2 | nc | 53.9 | 27.7 | 6 | yes |
| DUMMY3 | nc | 30.9 | 42.5 | 3 | no |
| DUMMY4 | nc | 50.3 | 19.3 | 7 | no |
| DUMMY5 | nc | 42.8 | 38.8 | 4 | no |
| DUMMY6 | nc | 36.9 | 46.7 | 4 | yes |
visit1
visit1群体的SCFA的数据分布情况
df_v1 %>% dplyr::select(acetate_v1, propionate_v1, butyrate_v1, valerate_v1) %>%pastecs::stat.desc() %>% kable() %>% kable_styling()
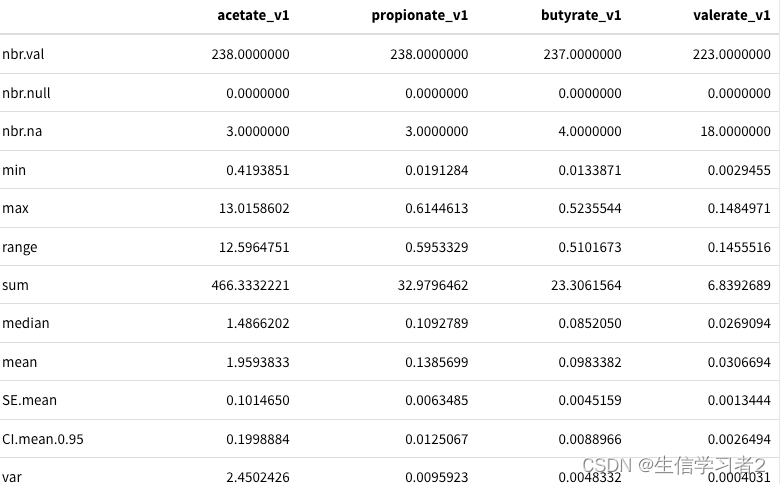
结果:不同的SCFA的数据描述结果,比如acetate_v1最大和最小值都要高于其他的。
visit2
visit2群体的SCFA的数据分布情况
df_v1 %>% dplyr::select(acetate_v2, propionate_v2, butyrate_v2, valerate_v2) %>%pastecs::stat.desc() %>% kable() %>% kable_styling()
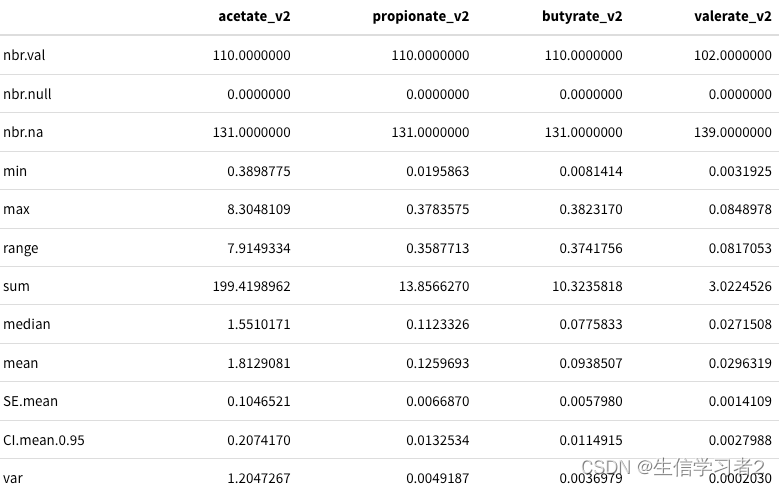
结果:不同的SCFA的数据描述结果,比如acetate_v1最大和最小值都要高于其他的。
SCFA ~ groups: box plots
线性回归校正abx_use因素比较不同分组的差异
plot_box <- function(group_lab, group_var, key_var, title, y.position, step.increase) {df_fig <- df_v1 %>%transmute(group = get(group_var), value = get(key_var), abx_use)df_fig$group <- factor(df_fig$group, ordered = FALSE)lm_fit <- lm(value ~ group + abx_use, data = df_fig)df_p <- data.frame(group1 = group_lab[1],group2 = group_lab[-1],p = summary(lm_fit)$coef[grepl("group", names(coef(lm_fit))), "Pr(>|t|)"]) %>%mutate(p = round(p, 2))bxp <- ggboxplot(df_fig, x = "group", y = "value", color = "group",add = "jitter", xlab = FALSE, ylab = FALSE, title = title) +stat_pvalue_manual(df_p, y.position = y.position, step.increase = step.increase, label = "p") +scale_color_npg(name = "Group") +theme(axis.text.x = element_blank(),axis.ticks.x = element_blank(),strip.background = element_rect(fill = "white"),legend.position = "bottom",plot.title = element_text(hjust = 0.5))return(bxp)
}
Primary group
Primary group: 按照频率分组
-
G1: # receptive anal intercourse = 0
-
G2: # receptive anal intercourse = 1
-
G3: # receptive anal intercourse = 2 - 5
-
G4: # receptive anal intercourse = 6 +
bxp_list <- list()
var_list <- c("acetate_v1", "butyrate_v1", "propionate_v1", "valerate_v1")
title_list <- c("Acetate", "Butyrate", "Propionate", "Valerate")
y_pos_list <- c(6, 0.25, 0.35, 0.07)
step_list <- c(0.1, 0.1, 0.1, 0.1)for (i in seq_along(var_list)) {bxp <- plot_box(group_lab = c("G1", "G2", "G3", "G4"),group_var = "group1",key_var = var_list[i], title = title_list[i],y.position = y_pos_list[i], step.increase = step_list[i])bxp_list[[i]] <- bxp
}ggarrange(bxp_list[[1]], bxp_list[[2]], bxp_list[[3]], bxp_list[[4]],common.legend = TRUE)
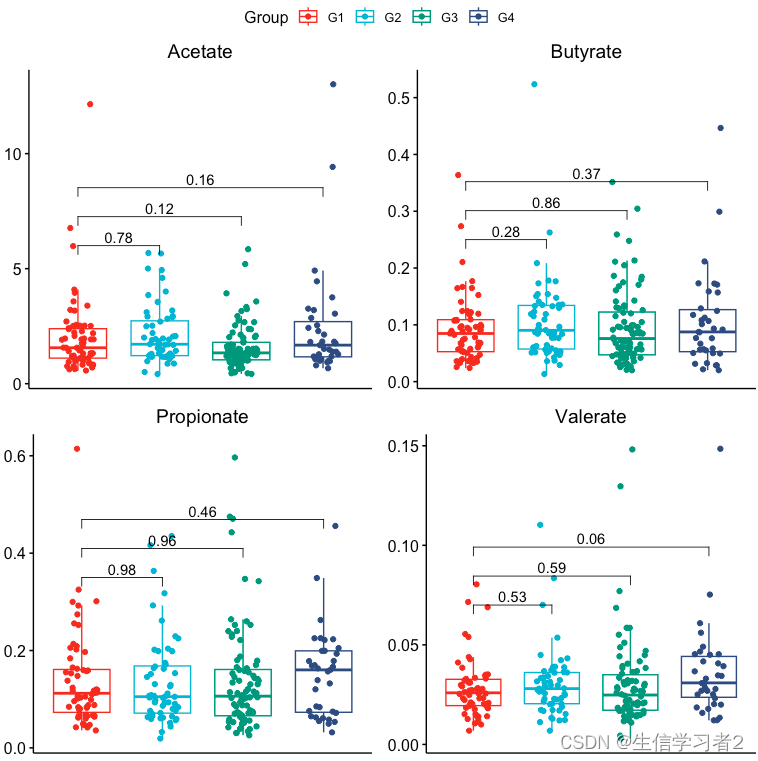
结果:上述短链脂肪酸在分组间均不存在显著差异。
Secondary group
Secondary group: 按照频率分组2
-
G1: # receptive anal intercourse = 0
-
G2: # receptive anal intercourse = 1
-
G3: # receptive anal intercourse = 2 - 3
-
G4: # receptive anal intercourse = 4 - 8
-
G5: # receptive anal intercourse = 9 +
bxp_list <- list()
var_list <- c("acetate_v1", "butyrate_v1", "propionate_v1", "valerate_v1")
title_list <- c("Acetate", "Butyrate", "Propionate", "Valerate")
y_pos_list <- c(6, 0.25, 0.35, 0.07)
step_list <- c(0.1, 0.1, 0.1, 0.1)for (i in seq_along(var_list)) {bxp <- plot_box(group_lab = c("G1", "G2", "G3", "G4", "G5"),group_var = "group2",key_var = var_list[i], title = title_list[i],y.position = y_pos_list[i], step.increase = step_list[i])bxp_list[[i]] <- bxp
}ggarrange(bxp_list[[1]], bxp_list[[2]], bxp_list[[3]], bxp_list[[4]],common.legend = TRUE)
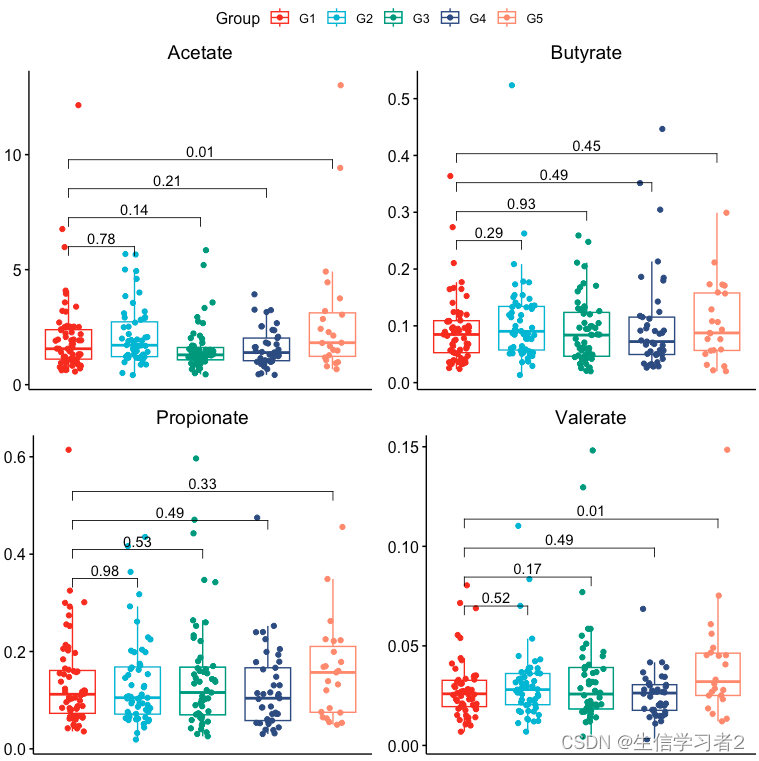
结果:短链脂肪酸在分组间的分布。
-
Acetate在G1和G5间显著差异; -
Valerate在G1和G5间显著差异
SCFA ~ groups: increasing trend
约束线性混合效应模型(CLME, Constrained Linear Mixed Effects Models):用于评估性暴露组和血浆炎症细胞因子水平之间的单调递增趋势,同时校正细菌抗生素使用情况。
plot_clme用于绘制趋势图
plot_clme <- function(clme_obj, group, y_min, y_max, p_gap, ann_pos, trend = "increase", ...) {overall_p <- clme_obj$p.valueind_p <- clme_obj$p.value.indest_mean <- clme_obj$theta[group]est_se <- sqrt(diag(clme_obj$cov.theta))[group]df_fig <- data.frame(x = group, y = est_mean, err = est_se)if (est_mean[2] < est_mean[length(est_mean)]) {start_p_pos <- est_mean[2] + p_gapend_p_pos <- max(est_mean) + p_gap} else if (est_mean[2] > est_mean[length(est_mean)]) {start_p_pos <- max(est_mean) + p_gapend_p_pos <- min(est_mean) + p_gap} else {if (trend == "increase") {start_p_pos <- est_mean[2] + p_gapend_p_pos <- max(est_mean) + 2 * p_gap} else {start_p_pos <- max(est_mean) + 2 * p_gapend_p_pos <- min(est_mean) + p_gap}}df_p <- data.frame(group1 = group[seq_len(length(group) - 1)],group2 = group[-1],x = group[-1],label = paste0("p = ", round(ind_p, 3)),y.position = seq.int(from = start_p_pos, to = end_p_pos, length.out = length(group) - 1))df_ann <- data.frame(x = group[1], y = ann_pos,fill = "white",label = paste0("Trend p-value = ", round(overall_p, 3)))fig <- df_fig %>%ggplot(aes(x = x, y = y)) +geom_bar(stat = "identity", color = "black", aes(fill = x)) + geom_errorbar(aes(ymin = y - 1.96 * err, ymax = y + 1.96 * err), width = .2, position = position_dodge(.9)) +add_pvalue(df_p,xmin = "group1",xmax = "group2",label = "label",y.position = "y.position",remove.bracket = FALSE, ...) +geom_label(data = df_ann, aes(x = x, y = y, label = label), size = 4, vjust = -0.5, hjust = 0, color = "black") +ylim(y_min, y_max) +theme_bw()return(fig)
}
-
P-value is obtained by linear regression adjusting for antibiotics usage.
-
P-values were not adjusted for multiple comparisons.
限制参数
cons <- list(order = "simple", decreasing = FALSE, node = 1)
Primary grouping: Acetate
短链脂肪酸Acetate与Primary grouping的相关关系
fit1 <- clme(acetate_v1 ~ group1 + abx_use, data = df_v1, constraints = cons, seed = 123)
summ_fit1 <- summary(fit1)fig_ace_primary <- plot_clme(summ_fit1, group = c("G1", "G2", "G3", "G4"), y_min = 0, y_max = 4, p_gap = 0.7, ann_pos = 3.5) +scale_fill_npg(name = NULL) +labs(x = NULL, y = "Acetate")fig_ace_primary
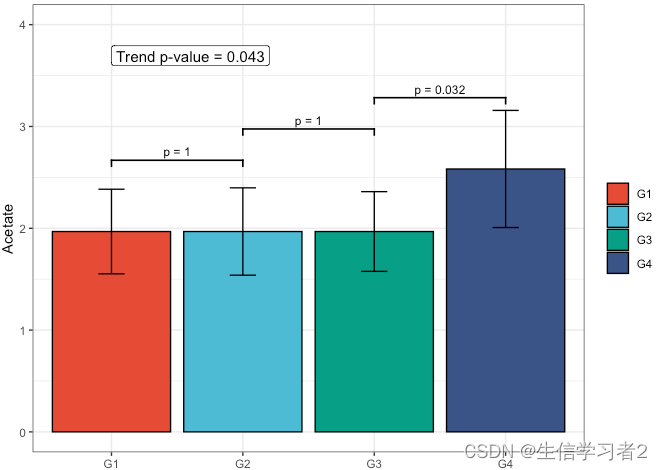
结果:acetate在不同组有显著差异的单调递增的趋势(G3和G4组存在显著差异)。
Primary grouping: Butyrate
短链脂肪酸Butyrate与Primary grouping的相关关系
fit2 <- clme(butyrate_v1 ~ group1 + abx_use, data = df_v1, constraints = cons, seed = 123)
summ_fit2 <- summary(fit2)fig_but_primary <- plot_clme(summ_fit2, group = c("G1", "G2", "G3", "G4"), y_min = 0, y_max = 0.18, p_gap = 0.04, ann_pos = 0.16) +scale_fill_npg(name = NULL) +labs(x = NULL, y = "Butyrate")fig_but_primary
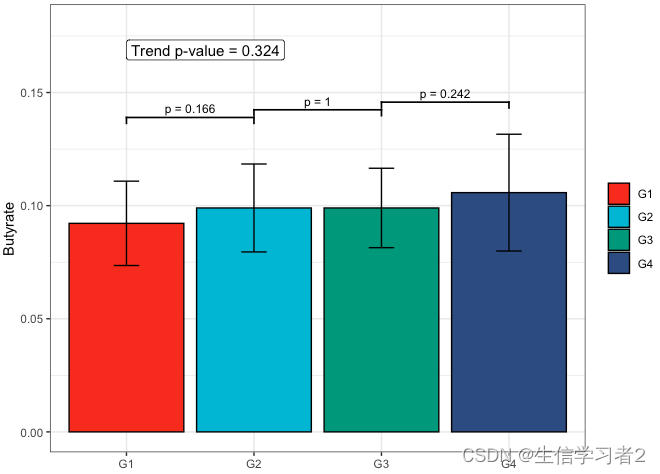
Primary grouping: Propionate
短链脂肪酸Propionate与Primary grouping的相关关系
fit3 <- clme(propionate_v1 ~ group1 + abx_use, data = df_v1, constraints = cons, seed = 123)
summ_fit3 <- summary(fit3)fig_pro_primary <- plot_clme(summ_fit3, group = c("G1", "G2", "G3", "G4"),y_min = 0, y_max = 0.25, p_gap = 0.05, ann_pos = 0.22) +scale_fill_npg(name = NULL) +labs(x = NULL, y = "Propionate")fig_pro_primary
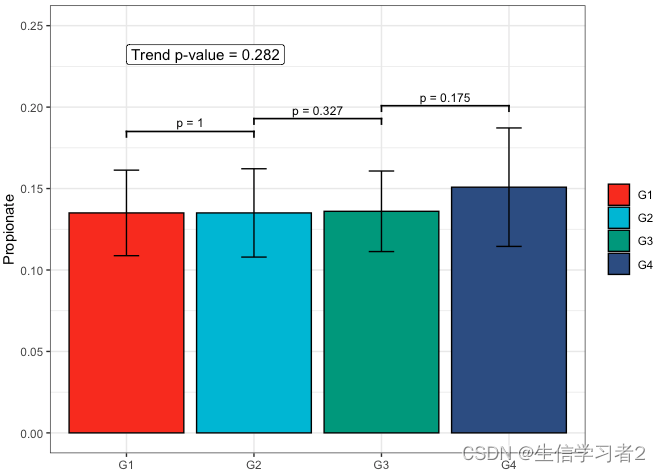
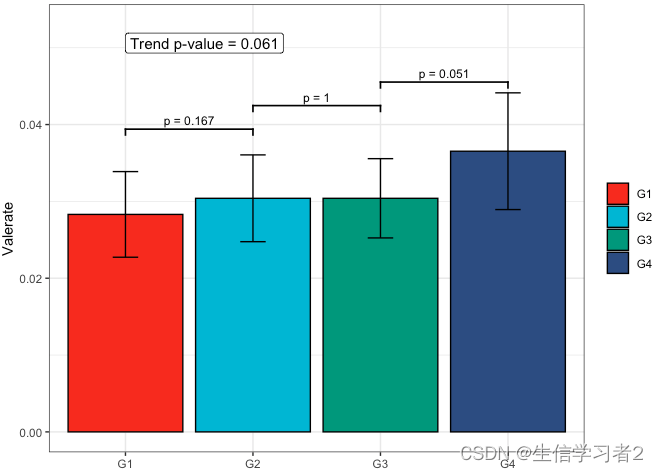
Primary grouping: Valerate
短链脂肪酸Valerate与Primary grouping的相关关系
fit4 <- clme(valerate_v1 ~ group1 + abx_use, data = df_v1, constraints = cons, seed = 123)
summ_fit4 <- summary(fit4)fig_val_primary <- plot_clme(summ_fit4, group = c("G1", "G2", "G3", "G4"),y_min = 0, y_max = 0.053, p_gap = 0.009, ann_pos = 0.048) +scale_fill_npg(name = NULL) +labs(x = NULL, y = "Valerate")fig_val_primary
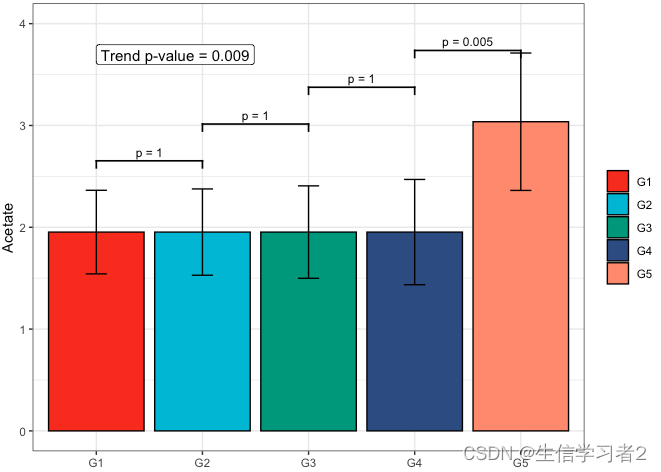
Secondary grouping: Acetate
短链脂肪酸Acetate与Secondary grouping的相关关系
fit1 <- clme(acetate_v1 ~ group2 + abx_use, data = df_v1, constraints = cons, seed = 123)
summ_fit1 <- summary(fit1)fig_ace_secondary <- plot_clme(summ_fit1, group = c("G1", "G2", "G3", "G4", "G5"), y_min = 0, y_max = 4, p_gap = 0.7, ann_pos = 3.5) +scale_fill_npg(name = NULL) +labs(x = NULL, y = "Acetate")fig_ace_secondary
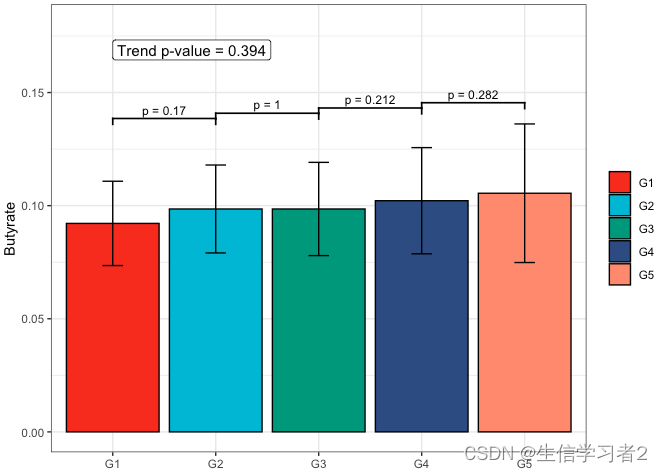
Secondary grouping: Butyrate
短链脂肪酸Butyrate与Secondary grouping的相关关系
fit2 <- clme(butyrate_v1 ~ group2 + abx_use, data = df_v1, constraints = cons, seed = 123)
summ_fit2 <- summary(fit2)fig_but_secondary <- plot_clme(summ_fit2, group = c("G1", "G2", "G3", "G4", "G5"), y_min = 0, y_max = 0.18, p_gap = 0.04, ann_pos = 0.16) +scale_fill_npg(name = NULL) +labs(x = NULL, y = "Butyrate")fig_but_secondary
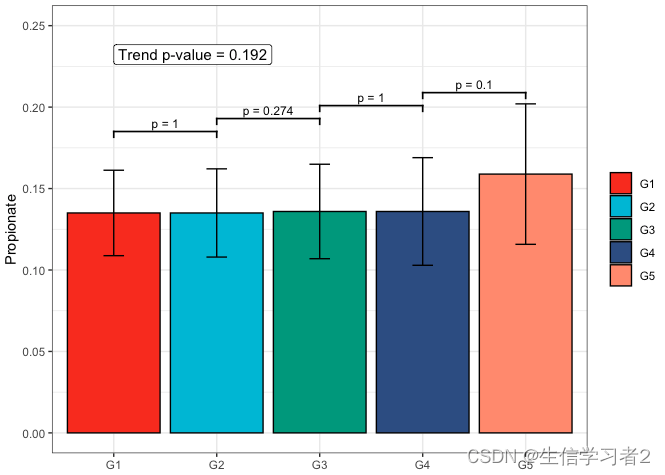
Secondary grouping: Propionate
短链脂肪酸Propionate与Secondary grouping的相关关系
fit3 <- clme(propionate_v1 ~ group2 + abx_use, data = df_v1, constraints = cons, seed = 123)
summ_fit3 <- summary(fit3)fig_pro_secondary <- plot_clme(summ_fit3, group = c("G1", "G2", "G3", "G4", "G5"),y_min = 0, y_max = 0.25, p_gap = 0.05, ann_pos = 0.22) +scale_fill_npg(name = NULL) +labs(x = NULL, y = "Propionate")fig_pro_secondary
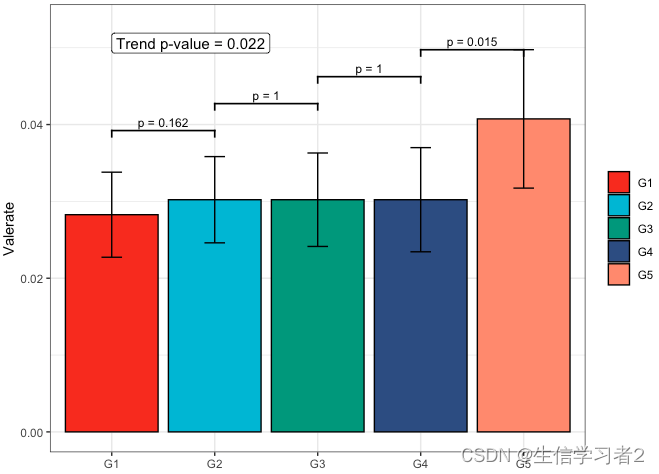
Secondary grouping: Valerate
短链脂肪酸Valerate与Secondary grouping的相关关系
fit4 <- clme(valerate_v1 ~ group2 + abx_use, data = df_v1, constraints = cons, seed = 123)
summ_fit4 <- summary(fit4)fig_val_secondary <- plot_clme(summ_fit4, group = c("G1", "G2", "G3", "G4", "G5"),y_min = 0, y_max = 0.053, p_gap = 0.009, ann_pos = 0.048) +scale_fill_npg(name = NULL) +labs(x = NULL, y = "Valerate")fig_val_secondary